Dear all,
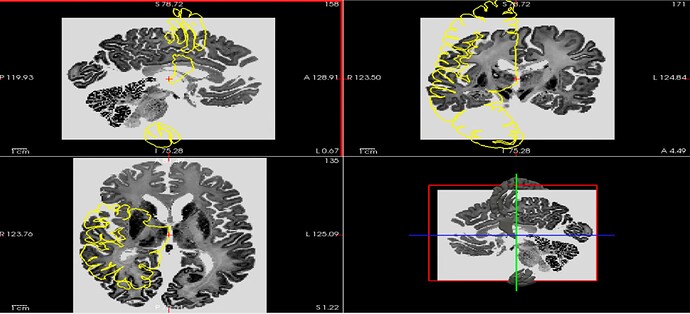
I am new to the community, and I am a researcher working on morphological brain complexity estimation. I want to use the amazing resources within the BigBrain project, but I have problems visualizing cortical layer surfaces overlaid with volumes. Specifically, I have been trying to use 3DSlicer and Freeview to visualize “BigBrainRelease.2015/3D_Volumes/Histological_Space/nii/full8_1000um_optbal.nii.gz” with the surface “BigBrainRelease.2015/Layer_Segmentation/3D_Surfaces/PLoSBiology2020/stl/layer0_left_327680.stl.gz” overlaid. The volume and the surface should live in the histology space, but it doesn’t look like it (see the Freeview screenshot below). The same happens when I use the MR image in the histology space “BigBrainRelease.2015/3D_MRI/nii/mri_to_blk_aligned.nii.gz”.
Any suggestions are most welcome!
Best,
Chiara